Published online Aug 28, 2021. doi: 10.37126/aige.v2.i4.117
Peer-review started: May 22, 2021
First decision: June 18, 2021
Revised: June 20, 2021
Accepted: July 15, 2021
Article in press: July 15, 2021
Published online: August 28, 2021
Artificial intelligence based approaches, in particular deep learning, have achieved state-of-the-art performance in medical fields with increasing number of software systems being approved by both Europe and United States. This paper reviews their applications to early detection of oesophageal cancers with a focus on their advantages and pitfalls. The paper concludes with future recommendations towards the development of a real-time, clinical implementable, interpretable and robust diagnosis support systems.
Core Tip: Precancerous changes in the lining of the oesophagus are easily missed during endoscopy as these lesions usually grow flat with only subtle change in colour, surface pattern and microvessel structure. Many factors impair the quality of endoscopy and subsequently the early detection of oesophageal cancer. Artificial intelligence (AI) solutions provide independence from the skills and experience of the operator in lesion recognition. Recent developments have introduced promising AI systems that will support the clinician in recognising, delineating and classifying precancerous and early cancerous changes during the endoscopy of the oesophagus in real-time.
- Citation: Gao X, Braden B. Artificial intelligence in endoscopy: The challenges and future directions. Artif Intell Gastrointest Endosc 2021; 2(4): 117-126
- URL: https://www.wjgnet.com/2689-7164/full/v2/i4/117.htm
- DOI: https://dx.doi.org/10.37126/aige.v2.i4.117
AI is the artificial intelligence exhibited by computer machines, which is in opposition to the natural intelligence that is displayed by human being, including consciousness and emotionality. With the advances on both computer hardware and software technology, at present, we are able to model about 600 K neurons and their interlaced connections, leading to processing over 100 million parameters. Since the human brain contains about 100 billion neurons[1], there is still a long way to go to before AI models are close enough to a human brain. Hence machine learning (ML) techniques are developed to perform task specific modelling that is in part supervised by human. While this supervised ML process is transparent and understandable, the human’s ability to comprehend large amounts of parameters, e.g., in millions, is limited, from a calculation point of view. Hence the application areas are restricted by employing semi- or fully supervised ML approaches. More recently, propelled by the advances of computer hardware, including large memory and graphics processing unit (GPU), task specific learning by computer itself, i.e., deep learning (DL), is realised, forming one of the most promising AI branches under the ML umbrella.
DL first made the headline when DL based computer program, AlphaGo, won the competition when playing board game Go with human players[2]. Since then, it has shown that nearly all winners in major competitions apply DL led methodologies, achieving state-of-the-art (SOTA) performance in nearly every domain, including natural language translation and image segmentation and classification. For example, the competition organised by Kaggle on detection of diabetics based on retinopathy has been won by DL based approach by a large margin in comparison with the other methods. While DL oriented methods have become a mainstream choice of meth
Hence this paper aims to review the latest development of application of AI to endoscopy realm and is organised below. Section 2 details the SOTA DL techniques and their application to medical domains. Section 3 explores the challenges facing early detection of oesophageal diseases from endoscopy and current solutions of computer aided systems. Section 4 points out future directions in achieving accurate diagnosis of oesophageal diseases with summaries provided in conclusion.
DL neural networks refer to a class of computing algorithms that can learn a hierarchy of features by establishing high-level attributes from low-level ones. One of the most popular models remains the convolutional neural network (CNN)[3], which comprises several (deep) layers of processing involving learnable operators (both linear and non-linear), by automating the process of constructing discriminative information from learning hierarchies. In addition, recent advances in computer hardware technology (e.g., the GPU) have propagated the implementation of CNNs in studying images. Usually, training a DL system to perform a task, e.g., classification, employs an arch
Conventionally, training a DL model requires large datasets and substantial training time. For example, the pre-trained CNN classifier, AlexNet[4], is built upon 7 Layers, simulating 500000 (K) neurons with 60 million (M) parameters and 630 M connections, and trained on a subset (1.2 M with 1 K categories) of ImageNet with 15 M 2D images of 22 K categories, taking up 16 d on a CPU and 1.6 d on a GPU. Usually, more data will lead to more accurate systems. In the development of electric cars of Tesla’s Autopilot, the training takes place with more than 780 million miles[5] whereas for playing AlphaGo[6] game using a computer, the training employed more than 100 million games.
DL-oriented approaches have recently been applied to medical images in a range of domains and achieved SOTA results. Although some doubt on DL has been casted on the ‘black box’ status while training without the embedding of human’s knowledge in the middle stages (e.g., hidden layers) apart from the initial input of labelled datasets, the performance of AI-led approached has been widely recognised, which is evidenced by the approval of medical devices by authorities. Between the year 2015 and 2020, 124 (about 15%) medical devices (mainly software) that are AL/ML/DL-based have been approved in Europe with Conformité Européene -marked and United States Food and Drug administration agency[7], highlighting the importance of AI/ML to the medical field, including an imaging system that uses algorithms to give diagnostic information for skin cancer and a smart electrocardiogram device that estimates the probability of a heart attack[8]. Table 1 summaries the recent achievements of DL-oriented approaches in medical domains.
| Ref. | Medical domain | Tasks |
| Muehlematter et al[7] | Skin | Diagnosis of skin cancer |
| United States Food and Drug Administration[8] | Electrocardiogram | Detection of heart attack |
| Pereira et al[9] | Retinopathy | Detection of diabetics |
| Gao et al[10] | Pulmonary CT images | Detection of tuberculosis types and severity |
| Sharib et al[11], Ali et al[12] | Endoscopy | Detection of artefact. |
| Gao et al[14] | CT Brain images | Classification of Alzheimer’s disease |
| Gao et al[15,16] | Pulmonary CT images | Analysis of multi-drug resistance |
| Gao et al[13] | Ultrasound | Classification of 3D echocardiographic video images |
| Wang et al[19], Ouyang et al[20] | Chest CT | Diagnosis of COVID-19 |
| Oh et al[21], Wang et al[22], Waheed et al[23] | Chest X-Ray | Diagnosis of COVID-19 |
| Everson et al[33], Horie et al[34], Ghatwary et al[35], Ohmori et al[38] | Endoscopy | Still image based cancer detection for 2 classes (normal vs abnormal) |
| de Groof et al[32], Everson et al[35], He et al[41], Guo et al[42] | Endoscopy | Video detection of SCC in real time |
| Gao et al[44], Tomita et al[45] | Endoscopy | Explainable AI for early detection of SCC |
Recently, AI or more specific DL-based approaches have won a number of competitions including the Kaggle competition on detection of diabetic retinopathy, segmentation of brain tumors from MRI images[9], analysis of severity of tuberculosis (TB) from high resolution 3D CT images in Image CLEFmed Competition[10] and detection of endoscopic artefacts from endoscopy video images in EAD2019[11] and EAD2020[12].
While applying AI/ML/DL approaches in medical domain, there are several challenges in need of responding. Firstly, in the medical domain, the number of datasets is limited, usually in hundreds whereas in other application, e.g., self-driving cars, datasets are in millions. Secondly, images are in multiple dimensions ranging from 2D to 5D (e.g., a moving heart at a specific location). And thirdly, perhaps the most outstanding obstacle is that medical data present subtle changes between normal and abnormal demanding the developed systems to be more precise.
Hence progress has been made to allow additional measures to be taken into account in order to apply DL techniques in medical fields. For example, for classification of 3D echocardiographic video images[13], a fused CNN architecture is established to incorporate both unsupervised CNN and hand-crafted features. For classification of 3D CT brain images[14], integration of both 2D and 3D CNN networks is in place. In addition, patch-based DL technique is designed to analyse 3D CT images for classification of TB types and analysis of multiple drug resistance[15,16] to overcome the sparse presence of diseased regions (< 10%). Another way to address small dataset issue is to employ transfer ML technique that is frequently implemented whereby a model developed built upon one dataset (e.g., ImageNet) for a specific task is reused as a starting point for a model on a different task with completely different datasets [e.g., coronavirus disease 2019 (COVID-19) computed tomography (CT) images]. Subsequently, most currently developed learning systems commence with a pre-trained model, such as VGG16[17] that is pre-trained on ImageNet datasets to extract initial feature maps that are then retrained to fit the new datasets and new tasks[18], capitalising on the accuracy a pre-trained model sustaining whilst saving considerable training times.
More recently, these AI techniques have been applied to predict COVID-19 virus and have demonstrated significant performance. With regard to medical images for diagnosis of COVID-19, CT and chest X-ray (CXR) represent the most common imaging tools. For 3D CT images, attention-based DL networks have shown effectiveness in classifying COVID-19 from normal subjects[19,20]. In relation to CXR, patch-based CNN is applied to study chest x-ray images[21] and to differentiate discriminatory features of COVID-19. In addition, COVID-Net[22], one of the pioneer studies, classifies COVID-19 from normal and pneumonia diseases through the application of a tailored DL network. To overcome the shortage of datasets, a number of researchers[23] apply generative adversarial neural network (GAN) to augment data first and subsequently to classify COVID-19.
In this paper, the application of AI/ML/DL techniques is exploited to endoscopy video images.
The oesophagus is the muscular tube that carries food and liquids from mouth to the stomach. The symptoms of oesophageal disorders include chest or back pain or having trouble swallowing. The most common problem with the oesophagus is gastroesophageal reflux disease which occurs when stomach contents frequently leak back, or reflux, into the oesophagus. The acidity of the fluids can irritate the lining of the oesophagus. Treatment of these disorders depends on the problem. Some problems get better with over-the-counter medicines or changes in diet. Others may need prescribed medicines or surgery.
As the 8th most common cancer worldwide[24], one of the most serious problems with regard to oesophagus is oesophageal cancer that constitutes the 6th leading cause of cancer-related death[25]. The main cancer types include adenocarcinoma and squamous cell carcinoma cancer (SCC). Globally, about 87% of all oesophageal cancers are in the form of SCC. The highest incidence rates often take place in Asia, the Middle East and Africa[26,27]. Early oesophageal cancer usually does not cause symptoms. At later stage, the symptoms might include swallowing difficulty, weight loss or continuous cough. Diagnosis of oesophageal cancer relies on imaging test, an upper endoscopy, and a biopsy.
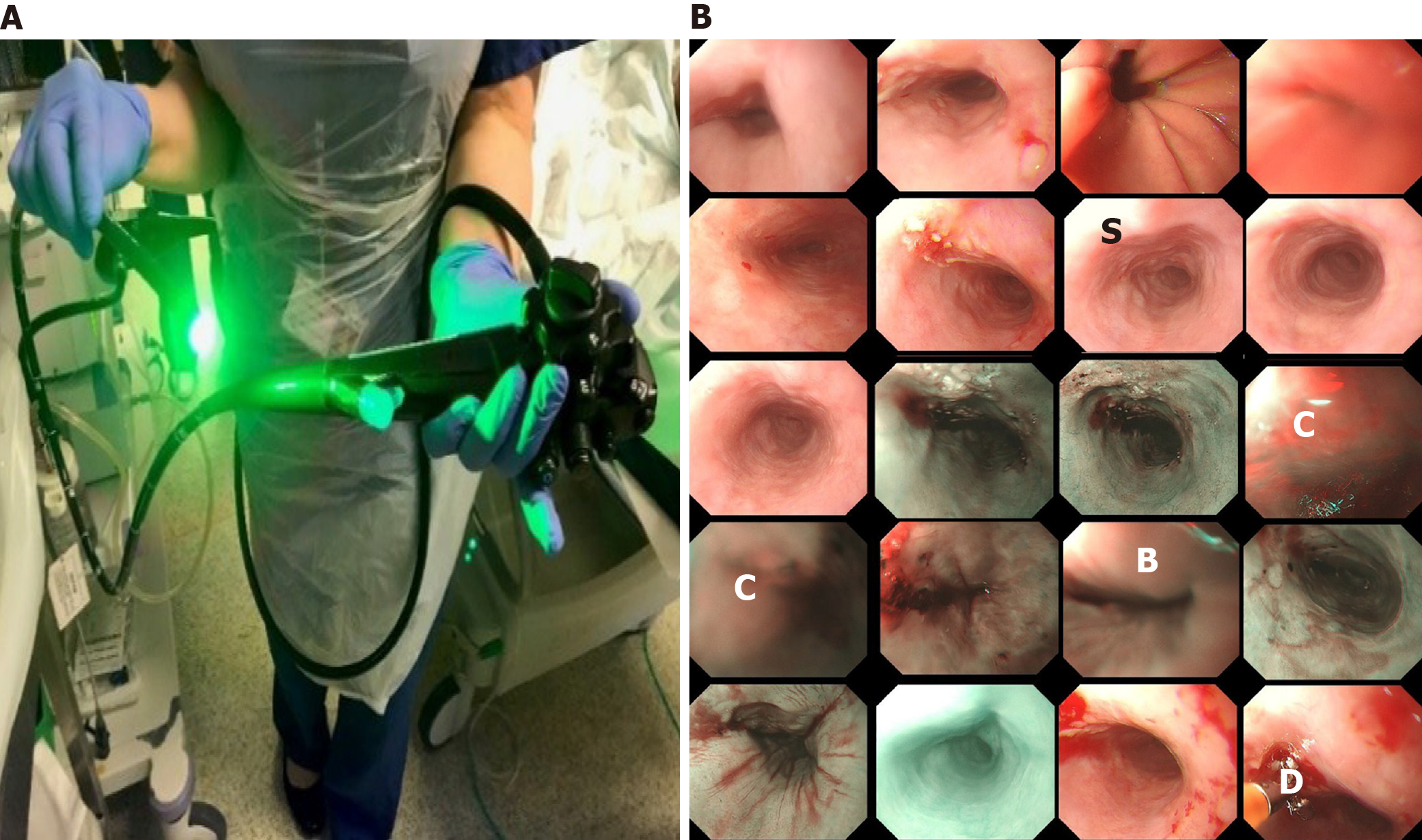
Optical endoscopy or endoscopy is the primary diagnostic and therapeutic tool for management of gastrointestinal malignancies, in particular oesophagus cancers. As illustrated in Figure 1A, to perform an endoscopy procedure of monitoring oesopha
While Figure 1 presents the surface of oesophageal walls, it also shows the artefact in a number of frames. This is because the movements of the inserted camera is confined within the limited space of the food pipe. The most common artefacts include colour misalignment (C), burry (B), saturation (S), and device (D) as demonstrated in Figure 1B.
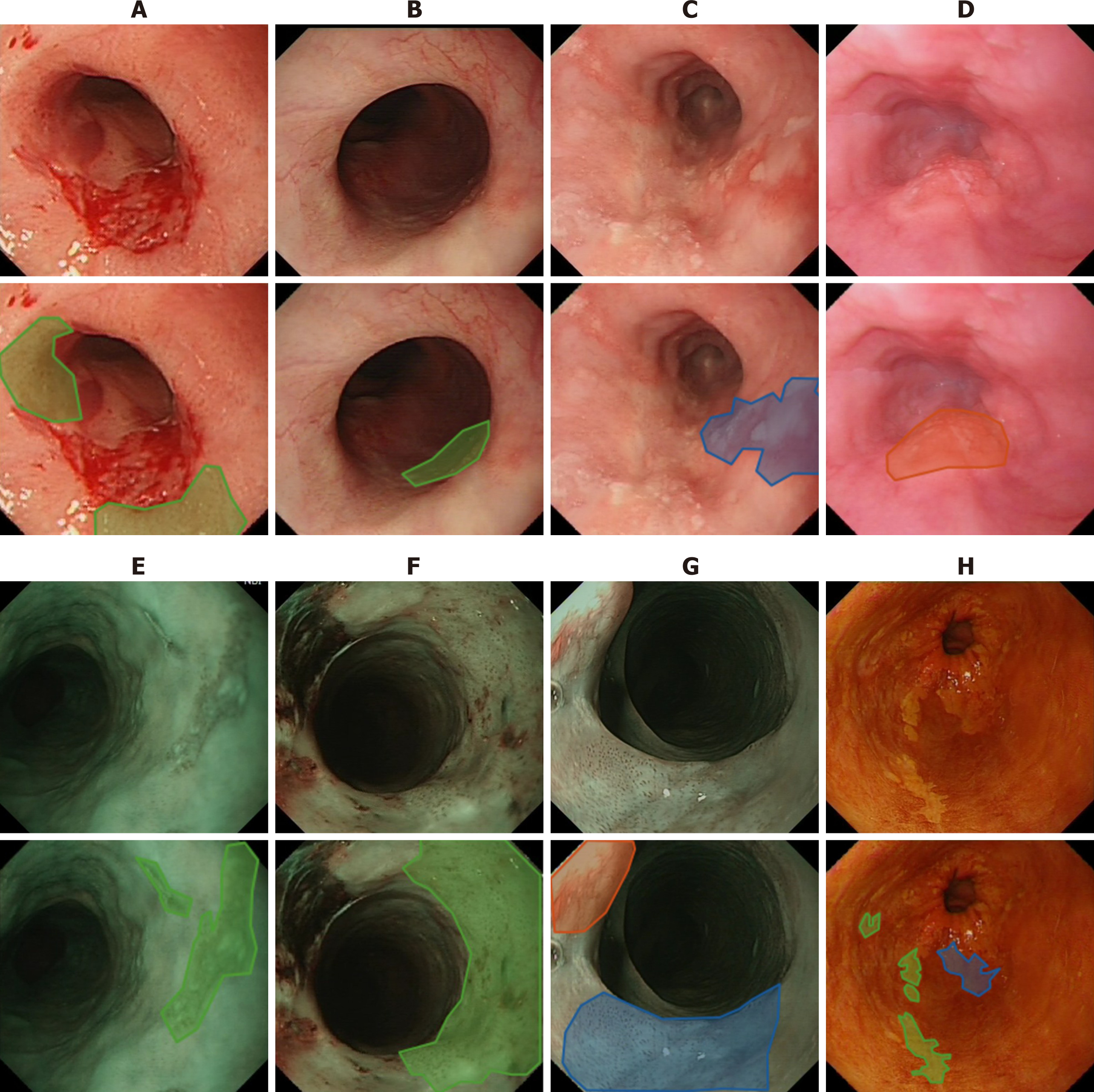
Commonly the five-year survival rate of oesophagus cancer is less than 20% as reported in[28]. However, this rate can be improved significantly to more than 90% if the cancer is detected in its early stages due to the fact that at this early stage, oesophageal cancer can be treated endoscopically[29], e.g., by removing diseased tissues or administrating (spraying) treatment drugs. The challenge lies here is that precancerous stages (dysplasia in the oesophageal squamous epithelium) and early stages of SCC display subtle changes in appearance (e.g., colour, surface structure) and in microvasculature, which therefore are easily missed at the time of conventional white light endoscopy (WLE) as illustrated in Figure 2A-D. To overcome this shortcoming while viewing WLE images, narrow-band imaging (NBI) can be turned on to display only two wavelengths [415 nm (blue) and 540 nm (green)] (Figure 2E-G) to improve the visibility of those suspected lesions by filtering out the rest of colour bands. Another approach is dye-based chromoendoscopy, i.e. Lugol’s staining technique, which highlights dysplastic abnormalities by spraying iodine[30] (Figure 2H).
While NBI technique improves the visibility of the vascular network and surface structure, it mainly facilitates the detection of unique vascular and pit pattern morphology that are present in neoplastic lesions[31], whereas precancerous stages can take a variety of forms. With the Lugol’s staining approach, many patients react uncomfortably to the spray.
It is therefore of clinical priority to have a computer assisted system to help clinicians to detect and highlight those potential suspected regions for further examinations. Currently, a number of promising results for computer-aided recognition of early neoplastic oesophageal lesions from endoscopic have been achieved based still images[32,33]. However, fewer less algorithms are applicable to real-time endoscopy to allow computer-aided decision-making during endoscopy at the point of examination. In addition, most of the existing studies focus mainly on the classification of endoscopic images between normal and abnormal stages with little work providing bounding boxes of the suspicious regions (detection) and delineating (segmentation).
Following challenges have been identified for the development of computerised algorithms for early detection of oesophageal cancers, which are inconspicuous changes on oesophageal surfaces artefacts of video images due to movement of endoscopic camera entering the food pipe limited time for patients undergoing each session of endoscopic procedure (about 20min) to minimise discomfort and invasiveness real time processing of video images to be in time to prompt endoscopist collecting biopsy samples while undertaking endoscopy limited datasets to train DL systems multiple modalities, including WLE, NBI and Lugol’s multiple classes, including LD, GD, SCC, normal, and artefact.
Progress on diagnosis of oesophageal cancer through the application of AI has been made by several research teams, mainly focusing on three directions, classification of abnormal from normal images, classification taking into consideration of processing speed, and detection of artefacts.
Horie et al[34] conducted research to distinguish oesophageal cancers from non-cancer patients with an aim to reproduce diagnostic accuracy. While applying conventional CNN architecture to classify two classes, the researchers have achieved 98% sensitivity for cancer detection. In the study conducted by Ghatwary et al[35], researchers have evaluated several SOTA CNN approaches aiming to achieve early detection of SCC from high-definition WLE (HD-WLE) images and come to the conclusion that the approaches of single shot detection[36] and Faster R-CNN[37] perform better. They use one image modality of WLE. Again, two classes are investigated in their study, i.e., cancerous and normal regions. While these studies demonstrate high accuracy of classification, the main focus of those research remains on the binary classification distinguishing abnormal from normal. Similarly, in the study by Ohmori et al[38], while the authors studied oesophageal lesions on several imaging modes including blue-laser images, only two classes of either cancer or non-cancer are classified by employing a deep neural network. For detection of any potential suspected regions regardless how small they are, segmentation of abnormal regions also plays a key role in supporting clinical decisions.
In addition, in order to assist clinicians in early diagnosis during endoscopic procedures, real-time processing of videos, i.e., with processing speed of 24+ frames per second (fps) or at most 41 milliseconds (ms) per frame, should be realised. Everson et al[33] have achieved inference time between 26 to 37ms for an image of 696 × 308 pixels. The work conducted by de Groof et al[32] requires 240ms to process each frame (i.e., 4.16 fps). For processing a video clip, frame processing and video playing back times all need to be considered to allow processed frames being played back seamlessly.
In order to ensure lesion detection takes place in time while patients undertaking endoscopy procedure, processing speed constitutes one of the key elements. Hence, comparisons are made to devalue the processing speed when detecting, classifying, and delineating multi-class (LD, HD, SCC) on multi-modality images (WLE, NBI, Lugol’s)[39] employing DL architectures of YOLOv3[40] and mask-CNN[41]. In this study by applying YOLOv3, the average processing time is in the range of 0.064-0.101 s per frame, which leads to 10-15 frames per second while processing frames of endoscopic videos with a resolution of 1920 × 1080 pixels. This work was conducted under Windows 10 operating system with 1 GPU (GeForce GTX 1060). The averaged accuracies for classification and detection can be realised to 85% and 74% respectively. Since YOLOv3 only provides bounding boxes without masks, the approach of mask-RCNN is utilised to delineate lesioned regions, producing classification, segmentation (masks) and bounding boxes. As a result, mask-RCNN achieves better detection result (i.e., bounding box) with 77% accuracy whereas the classification accuracy is similar to that obtained using YOLOYv3 with 84%. However, the processing speed applying mask-RCNN appears to be more than 10 times slower with an average of 1.2 s per frame, which is mainly stemmed from the time spent on the creation of masks. For the segmentation while employing mask-RCNN, the accuracy retains 63% measured on the overlapping regions between predicted and ground truth regions.
More recently, a research group by Guo et al[42] has developed a CAD system to aid decision making for early diagnosis of precancerous lesions. Their system can realise video processing time at 25 frames per second while applying narrow band images (NBI) that present clearer lesion structures than WLE. It appears that only one detection is identified for each frame, hence the study does not support localisation by bounding boxes.
Due to the confined space to film the oesophageal tube, a number of artefacts are present, which not only hamper clinician’s visual interpretation but also mislead training AI-based systems. Therefore, endoscopic artefact detection challenges were organised in 2019 (EAD2019)[11] and 2020 (EAD2020)[12] aiming to find solutions to these challenges. As expected, all top performant teams apply DL-based approaches to detect (bounding box), classify and segment artefacts including bubbles, saturation, blurry and artefacts[43].
While significant progress has been made towards development of AI-enhanced systems to support clinicians’ diagnosis, especially for early detection of oesophageal cancer, there is a still a considerable distance to go to benefit clinical diagnosis and to equip these assistant systems in an operative room. The following recommendations might shed light on future research directions.
Firstly, detection should be based on multi-classes, especially early onset lesions should be included. This is because most of the currently developed systems work on binary classifications between cancer and normal whereas cancers present most distinguishable visual features. At present, in 1 in 4 patients, the diagnosis of early stage oesophageal cancer is missed in their first visit[30]. Hence more work should emphasis on the detection of early onset of SCC. Only in this way can patients’ 5-year survival rates be increased to 90% from current 20%.
In addition, to circumvent data shortage, conventional data augmentation techniques appear to increase system accuracy by cropping, colour shifting, resizing and rotating. Due to the subtle change of early stages of SCC, data augmentation by inclusion of fake datasets generated by employing generative adversarial DL networks (GAN) appear to decrease the performance in this regard. Furthermore, when training with data that include samples with artefact, data augmentation with colour shifting also tend to hamper the system performance. Computational spectral imaging appears to benefit in this regard.
Secondly, to increase the wide acceptance by clinicians, the developed systems should be explainable and interpretable to a certain degree. For example, case-based reasoning[44] or attention-based modelling[45] are a way forward.
Lastly, real-time process should be achieved before the developed systems can make any real impact. This is because a collection of biopsy takes place only during the time of endoscopy. If those suspicious regions are overlooked, the patients in concern will miss the chances of correct diagnosis and appropriate treatment.
In conclusion, this paper overviews the current development of AI-based computer assisted systems for supporting early diagnosis of oesophageal cancers and proposes several future directions, expediting the clinical implementation and hence benefiting both patient and clinician communities.
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: United Kingdom
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Muneer A S-Editor: Liu M L-Editor: A P-Editor: Zhang YL
| 1. | von Bartheld CS, Bahney J, Herculano-Houzel S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J Comp Neurol. 2016;524:3865-3895. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 625] [Cited by in F6Publishing: 540] [Article Influence: 67.5] [Reference Citation Analysis (0)] |
| 2. | Silver D, Huang A, Maddison CJ, Guez A, Sifre L, van den Driessche G, Schrittwieser J, Antonoglou I, Panneershelvam V, Lanctot M, Dieleman S, Grewe D, Nham J, Kalchbrenner N, Sutskever I, Lillicrap T, Leach M, Kavukcuoglu K, Graepel T, Hassabis D. Mastering the game of Go with deep neural networks and tree search. Nature. 2016;529:484-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6655] [Cited by in F6Publishing: 2433] [Article Influence: 304.1] [Reference Citation Analysis (1)] |
| 3. | LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436-444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 36149] [Cited by in F6Publishing: 17272] [Article Influence: 1919.1] [Reference Citation Analysis (0)] |
| 4. | Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60:84-90. [DOI] [Cited in This Article: ] |
| 5. | Bradley R, Tesla Auopilot. 2016 Feb 23 [cited 20 April 2021]. In: MIT Technology Review. [Internet]. Available from: https://www.technologyreview.com/technology/tesla-autopilot/. [Cited in This Article: ] |
| 6. |
Gibney E.
Self-taught AI is best yet at strategy game Go, |
| 7. | Muehlematter UJ, Daniore P, Vokinger KN. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015-20): a comparative analysis. Lancet Digit Health. 2021;3:e195-e203. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 190] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 8. | US Food and Drug Administration. Proposed regulatory framework for modifications to artificial intel-ligence/machine learning (AI/ML)-based software as a medical device (SaMD). 2019. [cited 5 February 2021]. [Internet]. https://www.fda.gov/media/122535/download. [Cited in This Article: ] |
| 9. | Pereira S, Pinto A, Alves V, Silva CA. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans Med Imaging. 2016;35:1240-1251. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1538] [Cited by in F6Publishing: 814] [Article Influence: 101.8] [Reference Citation Analysis (0)] |
| 10. | Gao X, James-Reynolds C, Currie E. Analysis TB severity with an enhanced deep residual learning depth-resnet. In: CLEF2018 Working Notes. CEUR Workshop Proceedings, Avignon, France. 2018. Available from: http://ceur-ws.org/Vol-2125/paper_175.pdf. [Cited in This Article: ] |
| 11. | Ali S, Zhou F, Braden B, Bailey A, Yang S, Cheng G, Zhang P, Li X, Kayser M, Soberanis-Mukul RD, Albarqouni S, Wang X, Wang C, Watanabe S, Oksuz I, Ning Q, Khan MA, Gao XW, Realdon S, Loshchenov M, Schnabel JA, East JE, Wagnieres G, Loschenov VB, Grisan E, Daul C, Blondel W, Rittscher J. An objective comparison of detection and segmentation algorithms for artefacts in clinical endoscopy. Sci Rep. 2020;10:2748. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 31] [Cited by in F6Publishing: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Ali S, Dmitrieva M, Ghatwary N, Bano S, Polat G, Temizel A, Krenzer A, Hekalo A, Guo YB, Matuszewski B, Gridach M, Voiculescu I, Yoganand V, Chavan A, Raj A, Nguyen NT, Tran DQ, Huynh LD, Boutry N, Rezvy S, Chen H, Choi YH, Subramanian A, Balasubramanian V, Gao XW, Hu H, Liao Y, Stoyanov D, Daul C, Realdon S, Cannizzaro R, Lamarque D, Tran-Nguyen T, Bailey A, Braden B, East JE, Rittscher J. Deep learning for detection and segmentation of artefact and disease instances in gastrointestinal endoscopy. Med Image Anal. 2021;70:102002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 66] [Cited by in F6Publishing: 35] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 13. | Gao X, Li W, Loomes M, Wang L. A fused deep learning architecture for viewpoint classification of echocardiography. Informat Fusion. 2017;36:103-113. [DOI] [Cited in This Article: ] |
| 14. | Gao XW, Hui R, Tian Z. Classification of CT brain images based on deep learning networks. Comput Methods Programs Biomed. 2017;138:49-56. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 213] [Cited by in F6Publishing: 127] [Article Influence: 18.1] [Reference Citation Analysis (0)] |
| 15. | Gao, X, Carl James-Reynolds, Edward Currie. Analysis of Tuberculosis Severity Levels From CT Pulmonary Images Based on Enhanced Residual Deep Learning Architecture. Neuro Computing. 2021;392: 233-244. [DOI] [Cited in This Article: ] [Cited by in Crossref: 26] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 16. | Gao X, Quan Y. Prediction of multi-drug resistant TB from CT pulmonary Images based on deep learning techniques. Mol Pharmaceutics. 2018;15:4326-4335. [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 17. | Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. 2014 Sep 4 [cited 20 April 2021]. In: arXiv [Internet]. Available from: https://arxiv.org/abs/1409.1556. [Cited in This Article: ] |
| 18. | Rezvy S, Zebin T, Braden B, Pang W, Taylor S, Gao X. Transfer Learning For Endoscopy Disease Detection & Segmentation With Mask-RCNN Benchmark Architecture. Proceedings of the 2nd Inter-national Workshop and Challenge on Computer Vision in Endoscopy (EndoCV2020). 3 Apr 2020. Available from: http://ceur-ws.org/Vol-2595/endoCV2020_paper_id_17.pdf. [Cited in This Article: ] |
| 19. | Wang J, Bao Y, Wen Y, Lu H, Luo H, Xiang Y, Li X, Liu C, Qian D. Prior-Attention Residual Learning for More Discriminative COVID-19 Screening in CT Images. IEEE Trans Med Imaging. 2020;39:2572-2583. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 20. | Ouyang X, Huo J, Xia L, Shan F, Liu J, Mo Z, Yan F, Ding Z, Yang Q, Song B, Shi F, Yuan H, Wei Y, Cao X, Gao Y, Wu D, Wang Q, Shen D. Dual-Sampling Attention Network for Diagnosis of COVID-19 From Community Acquired Pneumonia. IEEE Trans Med Imaging. 2020;39:2595-2605. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 238] [Cited by in F6Publishing: 175] [Article Influence: 43.8] [Reference Citation Analysis (0)] |
| 21. | Oh Y, Park S, Ye JC. Deep Learning COVID-19 Features on CXR Using Limited Training Data Sets. IEEE Trans Med Imaging. 2020;39:2688-2700. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 523] [Cited by in F6Publishing: 349] [Article Influence: 87.3] [Reference Citation Analysis (0)] |
| 22. |
Wang L, Lin Z, Wong A, COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images, Sci Rep 2020; 10: 19549 [DOI: 10.
Wang L, Lin Z, Wong A, COVID-Net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest Radiography Images, |
| 23. | Waheed A, Goyal M. , Gupta D, Khanna A, Al-Turjman F, Pinheiro PR. IEEE Access. 2020;8:91916-91923. [DOI] [Cited in This Article: ] [Cited by in Crossref: 508] [Cited by in F6Publishing: 238] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 24. | Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394-424. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53206] [Cited by in F6Publishing: 50873] [Article Influence: 8478.8] [Reference Citation Analysis (44)] |
| 25. | Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400-412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1629] [Cited by in F6Publishing: 1800] [Article Influence: 163.6] [Reference Citation Analysis (4)] |
| 26. | Arnold M, Laversanne M, Brown LM, Devesa SS, Bray F. Predicting the Future Burden of Esophageal Cancer by Histological Subtype: International Trends in Incidence up to 2030. Am J Gastroenterol. 2017;112:1247-1255. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 278] [Cited by in F6Publishing: 237] [Article Influence: 33.9] [Reference Citation Analysis (1)] |
| 27. | Arnold M, Soerjomataram I, Ferlay J, Forman D. Global incidence of oesophageal cancer by histological subtype in 2012. Gut. 2015;64:381-387. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 944] [Cited by in F6Publishing: 941] [Article Influence: 104.6] [Reference Citation Analysis (0)] |
| 28. | Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104-117. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1848] [Cited by in F6Publishing: 2025] [Article Influence: 202.5] [Reference Citation Analysis (0)] |
| 29. | Shimizu Y, Tsukagoshi H, Fujita M, Hosokawa M, Kato M, Asaka M. Long-term outcome after endoscopic mucosal resection in patients with esophageal squamous cell carcinoma invading the muscularis mucosae or deeper. Gastrointest Endosc. 56:387-390. [DOI] [Cited in This Article: ] |
| 30. | Trivedi PJ, Braden B. Indications, stains and techniques in chromoendoscopy. QJM. 2013;106:117-131. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 53] [Cited by in F6Publishing: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 31. | van der Sommen F, Zinger S, Curvers WL, Bisschops R, Pech O, Weusten BL, Bergman JJ, de With PH, Schoon EJ. Computer-aided detection of early neoplastic lesions in Barrett's esophagus. Endoscopy. 2016;48:617-624. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 110] [Article Influence: 13.8] [Reference Citation Analysis (1)] |
| 32. | de Groof AJ, Struyvenberg MR, van der Putten J, van der Sommen F, Fockens KN, Curvers WL, Zinger S, Pouw RE, Coron E, Baldaque-Silva F, Pech O, Weusten B, Meining A, Neuhaus H, Bisschops R, Dent J, Schoon EJ, de With PH, Bergman JJ. Deep-Learning System Detects Neoplasia in Patients With Barrett's Esophagus With Higher Accuracy Than Endoscopists in a Multistep Training and Validation Study With Benchmarking. Gastroenterology. 2020;158:915-929.e4. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 172] [Cited by in F6Publishing: 174] [Article Influence: 43.5] [Reference Citation Analysis (0)] |
| 33. | Everson M, Herrera L, Li W, Luengo IM, Ahmad O, Banks M, Magee C, Alzoubaidi D, Hsu HM, Graham D, Vercauteren T, Lovat L, Ourselin S, Kashin S, Wang HP, Wang WL, Haidry RJ. Artificial intelligence for the real-time classification of intrapapillary capillary loop patterns in the endoscopic diagnosis of early oesophageal squamous cell carcinoma: A proof-of-concept study. United European Gastroenterol J. 2019;7:297-306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 52] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 34. | Horie Y, Yoshio T, Aoyama K, Yoshimizu S, Horiuchi Y, Ishiyama A, Hirasawa T, Tsuchida T, Ozawa T, Ishihara S, Kumagai Y, Fujishiro M, Maetani I, Fujisaki J, Tada T. Diagnostic outcomes of esophageal cancer by artificial intelligence using convolutional neural networks. Gastrointest Endosc. 2019;89:25-32. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 240] [Cited by in F6Publishing: 220] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 35. | Ghatwary N, Zolgharni M, Ye X. Early esophageal adenocarcinoma detection using deep learning methods. Int J Comput Assist Radiol Surg. 2019;14:611-621. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 36. | W, Anguelov D, Erhan D, Szegedy C, Reed S, Fu CY, Berg AC. SSD: Single Shot MultiBox Detector. Eur Conf Comput Vision. 2016;21-37. [DOI] [Cited in This Article: ] [Cited by in Crossref: 5649] [Cited by in F6Publishing: 5919] [Article Influence: 739.9] [Reference Citation Analysis (1)] |
| 37. | Girshick R. Fast R-CNN. Int Conf Comput Vision. 2015;1440-1448. [DOI] [Cited in This Article: ] |
| 38. | Ohmori M, Ishihara R, Aoyama K, Nakagawa K, Iwagami H, Matsuura N, Shichijo S, Yamamoto K, Nagaike K, Nakahara M, Inoue T, Aoi K, Okada H, Tada T. Endoscopic detection and differentiation of esophageal lesions using a deep neural network. Gastrointest Endosc. 2020;91:301-309.e1. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 72] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 39. | Gao XW, Braden B, Taylor S, Pang W. Towards Real-Time Detection of Squamous Pre-Cancers from Oesophageal Endoscopic Videos. Int Conf Comput Vision. 2019;1606-1612. [DOI] [Cited in This Article: ] |
| 40. | Redmon J, Farhadi A. YOLOv3: An incremental improvement. 2018 Apr 8 [cited 20 April 2021]. In: arXiv [Internet]. Available from https://arxiv.org/abs/1804.02767. [Cited in This Article: ] |
| 41. | He K, Gkioxari G, Dollar P, Girshick R. Mask R-CNN. Int Conf Comput Vision. 2017;2980-2988. [DOI] [Cited in This Article: ] |
| 42. | Guo L, Xiao X, Wu C, Zeng X, Zhang Y, Du J, Bai S, Xie J, Zhang Z, Li Y, Wang X, Cheung O, Sharma M, Liu J, Hu B. Real-time automated diagnosis of precancerous lesions and early esophageal squamous cell carcinoma using a deep learning model (with videos). Gastrointest Endosc. 2020;91:41-51. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 108] [Cited by in F6Publishing: 111] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 43. | Bolya D, Zhou C, Xiao F, and Lee YJ. YOLACT: Real-time Instance Segmentation. Int Conf Comput Vision. 2019;9156-916. [DOI] [Cited in This Article: ] |
| 44. | Gao XW, Braden B, Zhang L, Taylor S, Pang W, Petridis M. Case-based Reasoning of a Deep Learning Network for Prediction of Early Stage of Oesophageal Cancer, UKCBR 2019, AI-2019, Cambridge. 2019. Available from: http://www.expertupdate.org/papers/20-1/2019_paper_1.pdf. [Cited in This Article: ] |
| 45. | Tomita N, Abdollahi B, Wei J, Ren B, Suriawinata A, Hassanpour S. Attention-Based Deep Neural Networks for Detection of Cancerous and Precancerous Esophagus Tissue on Histopathological Slides. JAMA Netw Open. 2019;2:e1914645. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 113] [Cited by in F6Publishing: 77] [Article Influence: 15.4] [Reference Citation Analysis (0)] |