-
PDF
- Split View
-
Views
-
Cite
Cite
J L Raisaro, Francesco Marino, Juan Troncoso-Pastoriza, Raphaelle Beau-Lejdstrom, Riccardo Bellazzi, Robert Murphy, Elmer V Bernstam, Henry Wang, Mauro Bucalo, Yong Chen, Assaf Gottlieb, Arif Harmanci, Miran Kim, Yejin Kim, Jeffrey Klann, Catherine Klersy, Bradley A Malin, Marie Méan, Fabian Prasser, Luigia Scudeller, Ali Torkamani, Julien Vaucher, Mamta Puppala, Stephen T C Wong, Milana Frenkel-Morgenstern, Hua Xu, Baba Maiyaki Musa, Abdulrazaq G Habib, Trevor Cohen, Adam Wilcox, Hamisu M Salihu, Heidi Sofia, Xiaoqian Jiang, J P Hubaux, SCOR: A secure international informatics infrastructure to investigate COVID-19, Journal of the American Medical Informatics Association, Volume 27, Issue 11, November 2020, Pages 1721–1726, https://doi.org/10.1093/jamia/ocaa172
Close - Share Icon Share
Abstract
Global pandemics call for large and diverse healthcare data to study various risk factors, treatment options, and disease progression patterns. Despite the enormous efforts of many large data consortium initiatives, scientific community still lacks a secure and privacy-preserving infrastructure to support auditable data sharing and facilitate automated and legally compliant federated analysis on an international scale. Existing health informatics systems do not incorporate the latest progress in modern security and federated machine learning algorithms, which are poised to offer solutions. An international group of passionate researchers came together with a joint mission to solve the problem with our finest models and tools. The SCOR Consortium has developed a ready-to-deploy secure infrastructure using world-class privacy and security technologies to reconcile the privacy/utility conflicts. We hope our effort will make a change and accelerate research in future pandemics with broad and diverse samples on an international scale.
MISSION
A major lesson that the coronavirus disease 2019 (COVID-19) pandemic has already taught the scientific community is that timely international data sharing and collaborative data analysis is absolutely vital to navigate through policy decisions that have life-or-death consequences. Some of the most pressing issues about COVID-19 infections require urgent sharing of high-quality data concerning, for example, risk factors that influence infection, prognosis, and predictions of drug response from phenotypic, genotypic, and epigenetic data.1 To generate or test scientific hypotheses, we need large-scale and well-characterized patient-level datasets to provide sufficient statistical power. Building and sharing massive datasets containing personal health information have numerous legal and ethical implications that hinder new discoveries and prevent the scientific community from assessing their validity.2 In this respect, the case of 2 COVID-19 related articles published by The Lancet3 and The New England Journal of Medicine4 serves as an example. When concerns were raised regarding the veracity of the data used to support the conclusions in these articles, the 2 prestigious journals requested access to the raw data to conduct independent reviews. However, the authors could not comply with such a request, as granting access to the data would have violated confidentiality requirements, and the 2 journals had no choice but to retract the articles.3,5 These instances reinforce the need for a robust privacy- and confidentiality-compliant data-processing and sharing system to address these challenges in the era of COVID-19 and future pandemics.
Numerous data-driven projects have been launched across the globe to combat COVID-19, as summarized below. Yet, there is a lack of systematic support to address 1 of the main impediments that prevent and delay broad and sustainable medical data sharing: privacy protection. To address privacy protection challenges, researchers make trade-offs on data utility. On the 1 hand, several data-sharing projects on COVID-19 are based on a decentralized approach, employing the computation of local statistics (sometimes obfuscated to hide small numbers) that are subsequently shared and aggregated through meta-analysis. However, case numbers may sometimes be too low in certain subpopulations and could be considered identifiable information, which can make it very challenging for hospitals to even share aggregated data. Additionally, the approach only offers limited results and often depends on voluntary local analyses with human-in-the-loop approval and execution. On the other hand, other projects aim to centralize patient-level data from COVID-19 at a single site and then perform the analysis. Yet, that approach does not easily scale to international collaborations due to the heterogeneity and potential incompatibility of the various legal frameworks. We believe that there are more effective and privacy-congruent solutions to deal with this long-standing challenge and that privacy-by-design technology should be developed and is recently available for deployment to address the utmost urgency of data sharing by reducing administrative and regulatory barriers driven by privacy and security concerns. With this goal in mind, we have established an international consortium for Secure Collective Research (SCOR)6 to deploy the next-generation distributed infrastructure and tools for secure data sharing, analysis, and mining while respecting patient privacy and maximizing data utility during global disease outbreaks like the current COVID-19 pandemic. The list of founding partners for this global initiative is provided in Supplementary Material S1.
SHORT- AND LONG-TERM GOALS
SCOR aims to achieve the following goals:
Short-term: establish a proof-of-concept decentralized and privacy-preserving analytics platform, taking advantage of world-class privacy technology for COVID-19 data supporting cohort exploration for assessing the feasibility of research study protocols and facilitating speedy patient cohort recruitment.
Long-term: build a distributed privacy-preserving and sustainable infrastructure for federated statistical and machine learning analysis to support multicenter clinical studies of the COVID-19 outbreak and future pandemics.
POSITIONING OF SCOR REGARDING OTHER SIMILAR INITIATIVES
SCOR is a new initiative that complements existing multicentric data-sharing efforts to face the COVID-19 pandemic. COVID-19 research moves rapidly with new initiatives announced daily. In Table 1 we summarize the major initiatives we are aware of (as of June 2020) and compare them to SCOR along the following axes:
Comparison of SCOR with similar data-sharing initiatives
| Initiative . | Type of analysis . | Data storage . | Scope . | Type of data transferred . | Data protection mechanism . | Level of automation . |
|---|---|---|---|---|---|---|
| 4CE | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| ACT Network | cohort exploration | decentralized | national (USA) | aggregate-level | local obfuscation | fully automated system (SHRINEa) |
| LEOSS | centralized analytics | centralized | international (only EU) | patient-level | anonymization | manual analysis |
| OHDSI | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| PCORNet CDRNs | meta-analysis | decentralized | national (USA) | aggregate-level | local obfuscation | manual analysis |
| N3C | centralized analytics | centralized | national (USA) | patient-level | anonymization | manual analysis |
| SCOR | cohort exploration and decentralized analytics | decentralized | international | aggregate-level | encryption & global obfuscation | fully automated system (MedCOa) |
| Initiative . | Type of analysis . | Data storage . | Scope . | Type of data transferred . | Data protection mechanism . | Level of automation . |
|---|---|---|---|---|---|---|
| 4CE | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| ACT Network | cohort exploration | decentralized | national (USA) | aggregate-level | local obfuscation | fully automated system (SHRINEa) |
| LEOSS | centralized analytics | centralized | international (only EU) | patient-level | anonymization | manual analysis |
| OHDSI | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| PCORNet CDRNs | meta-analysis | decentralized | national (USA) | aggregate-level | local obfuscation | manual analysis |
| N3C | centralized analytics | centralized | national (USA) | patient-level | anonymization | manual analysis |
| SCOR | cohort exploration and decentralized analytics | decentralized | international | aggregate-level | encryption & global obfuscation | fully automated system (MedCOa) |
Comparison of fully automated systems for COVID-19 data sharing is reported in Table 2 below.
Comparison of SCOR with similar data-sharing initiatives
| Initiative . | Type of analysis . | Data storage . | Scope . | Type of data transferred . | Data protection mechanism . | Level of automation . |
|---|---|---|---|---|---|---|
| 4CE | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| ACT Network | cohort exploration | decentralized | national (USA) | aggregate-level | local obfuscation | fully automated system (SHRINEa) |
| LEOSS | centralized analytics | centralized | international (only EU) | patient-level | anonymization | manual analysis |
| OHDSI | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| PCORNet CDRNs | meta-analysis | decentralized | national (USA) | aggregate-level | local obfuscation | manual analysis |
| N3C | centralized analytics | centralized | national (USA) | patient-level | anonymization | manual analysis |
| SCOR | cohort exploration and decentralized analytics | decentralized | international | aggregate-level | encryption & global obfuscation | fully automated system (MedCOa) |
| Initiative . | Type of analysis . | Data storage . | Scope . | Type of data transferred . | Data protection mechanism . | Level of automation . |
|---|---|---|---|---|---|---|
| 4CE | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| ACT Network | cohort exploration | decentralized | national (USA) | aggregate-level | local obfuscation | fully automated system (SHRINEa) |
| LEOSS | centralized analytics | centralized | international (only EU) | patient-level | anonymization | manual analysis |
| OHDSI | meta-analysis | decentralized | international | aggregate-level | local obfuscation | manual analysis |
| PCORNet CDRNs | meta-analysis | decentralized | national (USA) | aggregate-level | local obfuscation | manual analysis |
| N3C | centralized analytics | centralized | national (USA) | patient-level | anonymization | manual analysis |
| SCOR | cohort exploration and decentralized analytics | decentralized | international | aggregate-level | encryption & global obfuscation | fully automated system (MedCOa) |
Comparison of fully automated systems for COVID-19 data sharing is reported in Table 2 below.
Type of analyses (cohort exploration vs meta-analysis vs distributed analytics vs centralized analytics)
Data storage (centralized vs decentralized)
Scope (national vs international)
Type of data transferred (aggregate-level vs patient-level)
Data protection mechanism (local obfuscation, global obfuscation, encryption)
Level of automation (manual analysis, semi-automated analysis, fully automated system)
The approach proposed by SCOR is the only 1 that (i) provides operational continuity for the long run, as it relies on a fully automated software platform for distributed data sharing; (ii) has an international scope; and (iii) provides the best data privacy/utility trade-offs, as it enables both cohort exploration and distributed analytics under strong privacy guarantees. These guarantees are ensured by deploying encryption techniques for distributed secure information aggregation across sites, lowering the need for local obfuscation.
CLINICAL RESEARCH GOALS
The rapid spread of the COVID-19 epidemic globally has almost overwhelmed health systems worldwide and it has already claimed lives in the hundreds of thousands. Starting from Asia, followed by Europe and next by the rest of the world, the first wave is now decreasing. No treatment has yet been demonstrated to be unequivocally effective and the subpopulation stratification of disease risks is still lacking, with multiple facets of presentation and prognosis. In particular, the recognized initial respiratory signs, symptoms, and laboratory findings have extended to many other settings, including dermatology, neurology, and hematology. Hospitals around the world have set up COVID-19 registries to accumulate information on symptoms, laboratory, respiratory function, imaging, and treatment to understand the disease. Joining forces will increase the number of patients that can be analyzed to address the next wave of the pandemic. Data harmonization will be challenging but, ultimately, essential. Similarly, the proposed secure and distributed data analysis approach will overcome obstacles to information sharing which some institutions are often reluctant to do. The SCOR network will serve as a hub for bringing together clinical research groups based on shared interests.
To demonstrate the utility of the SCOR approach, we will develop and apply use case scenarios (Box 1) that require data aggregation across multiple sites as each site has only a narrow view of the required information. This partial view stems from the uniqueness of the population at each site and from the difference in research protocols applied at each site.
SCOR REQUIREMENTS AND EXISTING DATA-SHARING PLATFORMS
The aim of SCOR is to provide an ecosystem for privacy-preserving distributed data analysis, which addresses all the 5 dimensions of secure data management, as expressed in the Five Safes framework14 (safe projects, safe people, safe setting, safe data, safe outputs) while overcoming the loss of data utility typical of existing decentralized approaches based on study-level meta-analyses that rely on site-level (ie, local) obfuscation to protect patients’ privacy. We distinguish between (i) safes that must be addressed at the consortium level (ie, safes that are enacted by decisions taken by the SCOR board [representative members from each participating institution] to pursue the high-level consortium’s privacy and security goals); and (ii) safes that must be addressed at the platform level (ie, safes that are enacted by technical safeguards featured by the technological infrastructure of the SCOR analysis platform). More details about the rational and platform requirements are discussed in Supplementary Material S0.
Table 2 briefly summarizes the most widespread distributed medical data analytics platforms in terms of provided functionalities and protection mechanisms to ensure safe settings and safe output requirements. We focus our comparison on the public platforms as they allow for an in-depth analysis. Yet, there exist also proprietary/closed platforms such as TriNetX, InSite, and Clinerion that, to the best of our knowledge, only partially address the data protection requirements for the SCOR initiative.
Comparison between available medical distributed analysis platforms
| . | Functionalities . | Safe settings . | Safe output . | ||
|---|---|---|---|---|---|
| Platform . | Cohort exploration . | Distributed analytics . | Secure aggregation . | Local obfuscation . | Global obfuscation . |
| SHRINE | • | • | |||
| Medical Informatics Platform | • | • | |||
| DataShield | • | • | • | ||
| MedCo | • | • | • | • | • |
| . | Functionalities . | Safe settings . | Safe output . | ||
|---|---|---|---|---|---|
| Platform . | Cohort exploration . | Distributed analytics . | Secure aggregation . | Local obfuscation . | Global obfuscation . |
| SHRINE | • | • | |||
| Medical Informatics Platform | • | • | |||
| DataShield | • | • | • | ||
| MedCo | • | • | • | • | • |
Comparison between available medical distributed analysis platforms
| . | Functionalities . | Safe settings . | Safe output . | ||
|---|---|---|---|---|---|
| Platform . | Cohort exploration . | Distributed analytics . | Secure aggregation . | Local obfuscation . | Global obfuscation . |
| SHRINE | • | • | |||
| Medical Informatics Platform | • | • | |||
| DataShield | • | • | • | ||
| MedCo | • | • | • | • | • |
| . | Functionalities . | Safe settings . | Safe output . | ||
|---|---|---|---|---|---|
| Platform . | Cohort exploration . | Distributed analytics . | Secure aggregation . | Local obfuscation . | Global obfuscation . |
| SHRINE | • | • | |||
| Medical Informatics Platform | • | • | |||
| DataShield | • | • | • | ||
| MedCo | • | • | • | • | • |
PROPOSED PLATFORM: MEDCO
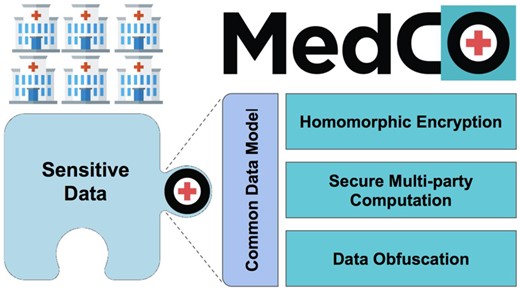
Given the SCOR platform requirements, the MedCo analysis platform15 is the 1 that best addresses them (Figure 1).
MedCo core technologies. MedCo is a decentralized software system that uses cutting-edge privacy-preserving technologies to enable the secure sharing of medical data among health institutions. It builds on 3 core privacy-preserving technologies: homomorphic encryption, secure multiparty computation, and data obfuscation. These technologies are used in synergy to combine information owned by multiple institutions and reveal otherwise hidden global insights while addressing legal and privacy concerns.
PRIVACY-PRESERVING TECHNOLOGICAL ENABLERS
Homomorphic encryption
Homomorphic encryption (HE)16 supports computation on encrypted data (ciphertexts). Thanks to this property, homomorphically encrypted data can be safely handed out to third parties who can perform meaningful operations on them without learning anything about their content. While fully homomorphic encryption schemes, (ie, schemes that enable arbitrary computations on ciphertexts) are still considered nonviable due to the high computational and storage overheads they introduce, practical schemes that enable only a limited number of computations on ciphertexts (eg, additions and multiplications) have reached a level of maturity that enables their use in real scenarios.
Secure multiparty computation
Secure multiparty computation (SMC)17 protocols allow multiple parties to jointly compute functions over their private inputs (eg, confidential patient-level data) without disclosing to the other parties more information about their inputs than what can be inferred from the output of the computation. This class of protocols is particularly attractive in privacy-preserving distributed analytic platforms due to the great variety of secure computations they enable. However, this flexibility often comes with a number of drawbacks that hinder their adoption, including high network overhead and the requirement of parties to be online during the computation. HE and SMC can be fruitfully employed in combination to mitigate their respective overheads and limitations and to provide effective solutions for privacy-preserving distributed analysis on sensitive data.
Data obfuscation
Data obfuscation techniques reduce the input data detail to an acceptable minimum and limit the information leakage stemming from the disclosure of the results. Indeed, even if data are kept private, the results of analyses performed may still reveal information about subjects that can be used to infer sensitive properties. Data obfuscation techniques alter data in a deterministic manner (eg, k-anonymity18 often applied to input data) or statistical manner (eg, differential privacy19 often implemented into processing methods to ensure safe outputs). For the results to remain useful, the amount of noise introduced by data obfuscation has to be carefully calibrated to reach the desired trade-off between utility and privacy. Studies show that k-anonymity and differential privacy sometimes give disappointing results when the target sample size is small.20,21 It is not a problem of both mechanisms but the unavoidable challenges in maneuvering statistics with limited flexibility. This issue is alleviated when safe settings are used to create large (protected) virtual datasets compared to applying data obfuscation to local datasets.
OPERATING PRINCIPLES
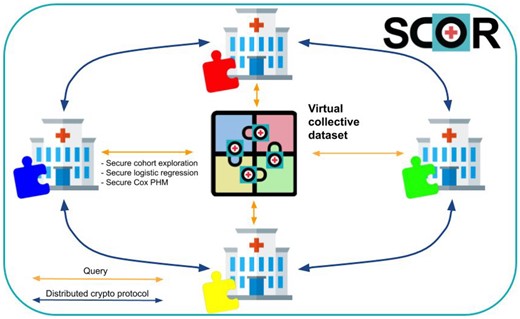
By using MedCo, health professionals and scientists can query data scattered among diverse institutions as if it were stored in a single location (virtual collective dataset) but without the need of seeing nor moving the data (see Figure 2). As such, it facilitates compliance with stringent data protection regulations such as the EU General Data Protection Regulation22 and the US Health Insurance Portability and Accountability Act.23 We include details about access control and accountability in Supplementary Material S7 and SCOR deployment plan in Supplementary Material S8.
The SCOR MedCo approach: when an institution queries the virtual collective dataset, it engages in a distributed cryptographic protocol with all the other institutions to securely obtain the result of the query. MedCo provides end-to-end protection against unauthorized access to data thanks to homomorphic encryption, which allows keeping the data in an encrypted state not only at rest and in transit but also during computation (safe settings). MedCo also removes the need for a central trusted authority by leveraging secure multiparty computation. The result of a query/analysis can be decrypted only through a distributed protocol that involves the approval of all the participating institutions. If 1 or more institutions are compromised by a cyber attack, the others can refuse to decrypt the data, thus keeping the data secure.
ETHICAL ISSUES
Ethical issues in data sharing and analysis are on the rise. Our technology provides privacy and security safeguards to automate global information exchange, but it might make the direct assessment of healthcare disparity harder due to the obfuscation. Fairness, equity, and transparency of medical informatics models represent the fundamental considerations for public trust and clinical usability. Many seemingly objective models are indeed influenced by their design, which can significantly over- or underestimate the risks on different subpopulations and introduce an unjustified basis for discriminating against a subpopulation. Such problems might be aggravated in a federated network with strong security protection and, if unnoticed, could result in significant ethical challenges. As a community, we should take a high standard in addressing these problems by design to consider fairness, equality, and justice to conduct responsible medical research.
CONCLUSION
There is an urgent need for data sharing and analysis in COVID-19, but we should not give up privacy in responsible research under pandemics. It is crucial to work together and build a robust and scalable infrastructure with state-of-the-art security and privacy technology to enable automated federated data analysis to accelerate scientific discoveries to combat the SARS-CoV-2 outbreak and future pandemics. We are fully committed to establishing this international consortium of collective data and a knowledge discovery network to support clinical research to answer important questions.
FUNDING
This research received no specific grant from any funding agency in the public, commercial or not-for-profit sectors. MF is supported by COVID-19 Data Science Institute (DSI), Bar-Ilan University (grant number 247017).
AUTHOR CONTRIBUTIONS
JR, JP, BM, AG, and XJ were responsible for the conception and design of the paper. JR, JL, XJ, EB, MF, AG, Troncoso-Pastoriza, MB, YC, BM, JF, and HS drafted the paper. RM, MP, SW, EB, MF, BM, AH, AW, MB, AG, AT, MM, JV, CK, LS, and HS acquired and contributed data, participated in the discussion, and edited/reviewed the manuscript. JF, BM, HX, FM, Troncoso-Pastoriza, MK, YC, HS, Prasser participated in the idea discussion and reviewed/edited/contributed to the manuscript. RB, RB, AH, YK, HW, TC reviewed the manuscript and conducted the final approval of the version to be published. All authors agreed to submit the report for publication.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
CONFLICT OF INTEREST STATEMENT
RB is a shareholder of Biomeris s.rl. HX have financial related interest at Melax Technologies Inc. RB serves as a Real World Evidence consultant for Pharmaceutical industry (UCB Pharma). The other co-authors have no competing interests to declare.
REFERENCES
Kaiser J.
Secure Covid Research | Secure Collective Covid-19 Research. https://securecovidresearch.org/ Accessed May 5, 2020
COVID19 | Cancer Genomics and BioComputing of Complex Diseases Lab. Cancer Genomics and BioComputing Lab;
MedCo | Collective protection of medical data. https://medco.epfl.ch/ Accessed April 13, 2020
General Data Protection Regulation (GDPR) Compliance Guidelines. GDPR.eu. https://gdpr.eu/ Accessed May 5, 2020
Health Insurance Portability and Accountability Act (HIPAA). http://www.hhs.gov/ocr/hipaa Accessed June 16, 2020.