Triclosan Alters Microbial Communities in Freshwater Microcosms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Water Collection & Microcosms
2.2. DNA Extraction and Sequencing
2.3. Processing and Visualization of 16S rRNA Sequence Data
2.4. qPCR of 16S rRNA and Int1 Genes
3. Results
3.1. Triclosan Decreases Richness and Evenness in Freshwater Microbial Communities
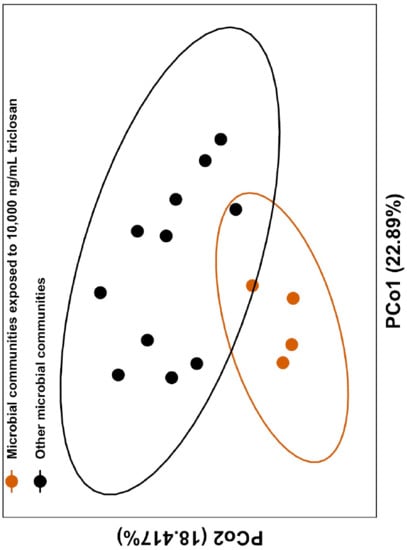
3.2. Triclosan Alters Freshwater Microbial Communities
3.3. Triclosan Addition Does Not Increase Int1 Abundance in Freshwater Microcosms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dhillon, G.S.; Kaur, S.; Pulicharla, R.; Brar, S.K.; Cledón, M.; Verma, M.; Surampalli, R.Y. Triclosan: Current status, occurrence, environmental risks and bioaccumulation potential. Int. J. Environ. Res. Public. Health 2015, 12, 5657–5684. [Google Scholar] [CrossRef] [PubMed]
- Chattopadhyay, D. Antibacterial consumer products: How reliable are they? Resonance 2017, 22, 761–767. [Google Scholar] [CrossRef]
- Kolpin, D.W.; Furlong, E.T.; Meyer, M.T.; Thurman, E.M.; Zaugg, S.D.; Barber, L.B.; Buxton, H.T. Pharmaceuticals, hormones, and other organic wastewater contaminants in U.S. streams, 1999–2000: A national reconnaissance. Environ. Sci. Technol. 2002, 36, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Drury, B.; Scott, J.; Rosi-Marshall, E.J.; Kelly, J.J. Triclosan exposure increases triclosan resistance and influences taxonomic composition of benthic bacterial communities. Environ. Sci. Technol. 2013, 47, 8923–8930. [Google Scholar] [CrossRef]
- Robertshaw, H.; Leppard, B. Contact dermatitis to triclosan in toothpaste. Contact Dermatitis 2007, 57, 383–384. [Google Scholar] [CrossRef]
- Yang, H.; Wang, W.; Romano, K.A.; Gu, M.; Sanidad, K.Z.; Kim, D.; Yang, J.; Schmidt, B.; Panigrahy, D.; Pei, R.; et al. A common antimicrobial additive increases colonic inflammation and colitis-associated colon tumorigenesis in mice. Sci. Transl. Med. 2018, 10, eaan4116. [Google Scholar] [CrossRef]
- Dayan, A.D. Risk assessment of triclosan [Irgasan®] in human breast milk. Food Chem. Toxicol. 2007, 45, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Allmyr, M.; Adolfsson-Erici, M.; McLachlan, M.S.; Sandborgh-Englund, G. Triclosan in plasma and milk from Swedish nursing mothers and their exposure via personal care products. Sci. Total Environ. 2006, 372, 87–93. [Google Scholar] [CrossRef]
- Bever, C.S.; Rand, A.A.; Nording, M.; Taft, D.; Kalanetra, K.M.; Mills, D.A.; Breck, M.A.; Smilowitz, J.T.; German, J.B.; Hammock, B.D. Effects of triclosan in breast milk on the infant fecal microbiome. Chemosphere 2018, 203, 467–473. [Google Scholar] [CrossRef] [PubMed]
- Veldhoen, N.; Skirrow, R.C.; Osachoff, H.; Wigmore, H.; Clapson, D.J.; Gunderson, M.P.; Van Aggelen, G.; Helbing, C.C. The bactericidal agent triclosan modulates thyroid hormone-associated gene expression and disrupts postembryonic anuran development. Aquat. Toxicol. 2006, 80, 217–227. [Google Scholar] [CrossRef]
- Capkin, E.; Ozcelep, T.; Kayis, S.; Altinok, I. Antimicrobial agents, triclosan, chloroxylenol, methylisothiazolinone and borax, used in cleaning had genotoxic and histopathologic effects on rainbow trout. Chemosphere 2017, 182, 720–729. [Google Scholar] [CrossRef]
- Foran, C.M.; Bennett, E.R.; Benson, W.H. Developmental evaluation of a potential non-steroidal estrogen: Triclosan. Mar. Environ. Res. 2000, 50, 153–156. [Google Scholar] [CrossRef]
- Falisse, E.; Voisin, A.-S.; Silvestre, F. Impacts of triclosan exposure on zebrafish early-life stage: Toxicity and acclimation mechanisms. Aquat. Toxicol. 2017, 189, 97–107. [Google Scholar] [CrossRef]
- Gaulke, C.A.; Barton, C.L.; Proffitt, S.; Tanguay, R.L.; Sharpton, T.J. Triclosan Exposure Is Associated with Rapid Restructuring of the Microbiome in Adult Zebrafish. PLoS ONE 2016, 11, e0154632. [Google Scholar] [CrossRef] [PubMed]
- Narrowe, A.B.; Albuthi-Lantz, M.; Smith, E.P.; Bower, K.J.; Roane, T.M.; Vajda, A.M.; Miller, C.S. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure. Microbiome 2015, 3. [Google Scholar] [CrossRef]
- Richmond, E.K.; Grace, M.R.; Kelly, J.J.; Reisinger, A.J.; Rosi, E.J.; Walters, D.M. Pharmaceuticals and personal care products (PPCPs) are ecological disrupting compounds (EcoDC). Elem Sci Anth 2017, 5. [Google Scholar] [CrossRef]
- Carey, D.E.; McNamara, P.J. The impact of triclosan on the spread of antibiotic resistance in the environment. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Nietch, C.T.; Quinlan, E.L.; Lazorchak, J.M.; Impellitteri, C.A.; Raikow, D.; Walters, D. Effects of a chronic lower range of triclosan exposure on a stream mesocosm community. Environ. Toxicol. Chem. 2013, 32, 2874–2887. [Google Scholar] [CrossRef] [PubMed]
- Chuanchuen, R.; Beinlich, K.; Hoang, T.T.; Becher, A.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. Cross-Resistance between Triclosan and Antibiotics inPseudomonas aeruginosa Is Mediated by Multidrug Efflux Pumps: Exposure of a Susceptible Mutant Strain to Triclosan Selects nfxB Mutants Overexpressing MexCD-OprJ. Antimicrob. Agents Chemother. 2001, 45, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Mcmurry, L.M.; Oethinger, M.; Levy, S.B. Overexpression of marA, soxS, or acrAB produces resistance to triclosan in laboratory and clinical strains of Escherichia coli. FEMS Microbiol. Lett. 1998, 166, 305–309. [Google Scholar] [CrossRef] [PubMed]
- Jutkina, J.; Marathe, N.P.; Flach, C.-F.; Larsson, D.G.J. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 2018, 616–617, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Butler, E.; Whelan, M.J.; Ritz, K.; Sakrabani, R.; van Egmond, R. Effects of triclosan on soil microbial respiration. Environ. Toxicol. Chem. 2011, 30, 360–366. [Google Scholar] [CrossRef]
- Zaayman, M.; Siggins, A.; Horne, D.; Lowe, H.; Horswell, J. Investigation of triclosan contamination on microbial biomass and other soil health indicators. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef] [PubMed]
- Sissons, C.H.; Wong, L.; Cutress, T.W. Inhibition by ethanol of the growth of biofilm and dispersed microcosm dental plaques. Arch. Oral Biol. 1996, 41, 27–34. [Google Scholar] [CrossRef]
- Wadhwani, T.; Desai, K.; Patel, D.; Lawani, D.; Bahaley, P.; Joshi, P.; Kothari, V. Effect of various solvents on bacterial growth in context of determining MIC of various antimicrobials. Internet J. Microbiol. 2009, 7, 1–8. [Google Scholar]
- Wilson, B.; Chen, R.F.; Cantwell, M.; Gontz, A.; Zhu, J.; Olsen, C.R. The partitioning of Triclosan between aqueous and particulate bound phases in the Hudson River Estuary. Mar. Pollut. Bull. 2009, 59, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Dahan, D.; Jude, B.A.; Lamendella, R.; Keesing, F.; Perron, G.G. Exposure to Arsenic Alters the Microbiome of Larval Zebrafish. Front. Microbial. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Pindling, S.; Azulai, D.; Zheng, B.; Dahan, D.; Perron, G.G. Dysbiosis and early mortality in zebrafish larvae exposed to subclinical concentrations of streptomycin. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.-L.; Jiang, X.; Wu, Q.L.; Zhou, N.-Y. Intragenomic heterogeneity of 16S rRNA genes causes overestimation of prokaryotic diversity. Appl. Environ. Microbial. 2013, 79, 5962–5969. [Google Scholar] [CrossRef]
- Schliep, K.P. Phangorn: Phylogenetic analysis in R. Bioinformatics 2011, 27, 592–593. [Google Scholar] [CrossRef]
- Wittebolle, L.; Marzorati, M.; Clement, L.; Balloi, A.; Daffonchio, D.; Heylen, K.; De Vos, P.; Verstraete, W.; Boon, N. Initial community evenness favours functionality under selective stress. Nature 2009, 458, 623–626. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbial. 2005, 71, 8228–8235. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Hamady, M.; Kelley, S.T.; Knight, R. Quantitative and Qualitative β Diversity Measures Lead to Different Insights into Factors That Structure Microbial Communities. Appl. Environ. Microbiol. 2007, 73, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘vegan’. 2015. Available online: http://vegan.r-forge.r-project.org (accessed on 5 May 2019).
- McMurdie, P.J.; Holmes, S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [PubMed]
- Gaze, W.H.; Zhang, L.; Abdouslam, N.A.; Hawkey, P.M.; Calvo-Bado, L.; Royle, J.; Brown, H.; Davis, S.; Kay, P.; Boxall, A.B.; et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J. 2011, 5, 1253–1261. [Google Scholar] [CrossRef] [Green Version]
- Leonard, A.F.C.; Singer, A.; Ukoumunne, O.C.; Gaze, W.H.; Garside, R. Is it safe to go back into the water? A systematic review and meta-analysis of the risk of acquiring infections from recreational exposure to seawater. Int. J. Epidemiol. 2018, 47, 572–586. [Google Scholar] [CrossRef]
- Leonard, A.F.C.; Zhang, L.; Balfour, A.J.; Garside, R.; Gaze, W.H. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ. Int. 2015, 82, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Perron, G.G.; Inglis, R.F.; Pennings, P.S.; Cobey, S. Fighting microbial drug resistance: A primer on the role of evolutionary biology in public health. Evol. Appl. 2015, 8, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Perron, G.G.; Gonzalez, A.; Buckling, A. Source-sink dynamics shape the evolution of antibiotic resistance and its pleiotropic fitness cost. Proc. Biol. Sci. 2007, 274, 2351–2356. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.-P.; Matthijs, S.; Colak, H.; Chablain, P.; Bilocq, F.; Van Eldere, J.; De Vos, D.; Zizi, M.; Triest, L.; Cornelis, P. Global Pseudomonas aeruginosa biodiversity as reflected in a Belgian river. Environ. Microbial. 2005, 7, 969–980. [Google Scholar] [CrossRef] [PubMed]
- De Vos, D.; Pirnay, J.-P.; Bilocq, F.; Jennes, S.; Verbeken, G.; Rose, T.; Keersebilck, E.; Bosmans, P.; Pieters, T.; Hing, M.; et al. Molecular Epidemiology and Clinical Impact of Acinetobacter calcoaceticus-baumannii Complex in a Belgian Burn Wound Center. PLoS ONE 2016, 11, e0156237. [Google Scholar] [CrossRef] [PubMed]
- Birošová, L.; Mikulášová, M. Development of triclosan and antibiotic resistance in Salmonella enterica serovar Typhimurium. J. Med. Microbiol. 2009, 58, 436–441. [Google Scholar] [CrossRef] [Green Version]
- Song, B.; Kerkhof, L.J.; Häggblom, M.M. Characterization of bacterial consortia capable of degrading 4-chlorobenzoate and 4-bromobenzoate under denitrifying conditions. FEMS Microbiol. Lett. 2002, 213, 183–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.-M.; Lu, P.-Z. Characterization of bacterial communities in sediments receiving various wastewater effluents with high-throughput sequencing analysis. Microb. Ecol. 2014, 67, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Buth, J.M.; Steen, P.O.; Sueper, C.; Blumentritt, D.; Vikesland, P.J.; Arnold, W.A.; McNeill, K. Dioxin photoproducts of triclosan and its chlorinated derivatives in sediment cores. Environ. Sci. Technol. 2010, 44, 4545–4551. [Google Scholar] [CrossRef] [PubMed]
- Aranami, K.; Readman, J.W. Photolytic degradation of triclosan in freshwater and seawater. Chemosphere 2007, 66, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Kodani, S.; Imoto, A.; Mitsutani, A.; Murakami, M. Isolation and identification of the antialgal compound, harmane (1-methyl-β-carboline), produced by the algicidal bacterium, Pseudomonas sp. K44-1. J. Appl. Phycol. 2002, 14, 109–114. [Google Scholar] [CrossRef]
- Wright, S.J.L.; Thompson, R.J. Bacillus volatiles antagonize cyanobacteria. FEMS Microbiol. Lett. 1985, 30, 263–267. [Google Scholar] [CrossRef]


| Treatment | Observed | Shannon | ||
|---|---|---|---|---|
| Full | No Chloroplast | Full | No Chloroplast | |
| Control | 161.33 (10.69) | 148.33 (9.45) | 4.18 (0.12) | 4.10 (0.12) |
| 1 ng/L | 152.50 (10.28) | 141.00 (43.49) | 3.96 (0.16) | 3.90 (0.15) |
| 10 ng/L | 180.50 (44.38) | 168.00 (39.75) | 3.98 (0.13) | 3.94 (0.13) |
| 10,000 ng/L | 108.50 (13.34) | 101.25 (10.14) | 3.53 (0.41) | 3.49 (0.42) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Clarke, A.; Azulai, D.; Dueker, M.E.; Vos, M.; Perron, G.G. Triclosan Alters Microbial Communities in Freshwater Microcosms. Water 2019, 11, 961. https://doi.org/10.3390/w11050961
Clarke A, Azulai D, Dueker ME, Vos M, Perron GG. Triclosan Alters Microbial Communities in Freshwater Microcosms. Water. 2019; 11(5):961. https://doi.org/10.3390/w11050961
Chicago/Turabian StyleClarke, Alexandra, Daniella Azulai, M. Elias Dueker, Michiel Vos, and Gabriel G. Perron. 2019. "Triclosan Alters Microbial Communities in Freshwater Microcosms" Water 11, no. 5: 961. https://doi.org/10.3390/w11050961