Detection of Infectious Noroviruses from Wastewater and Seawater Using PEMAXTM Treatment Combined with RT-qPCR
Abstract
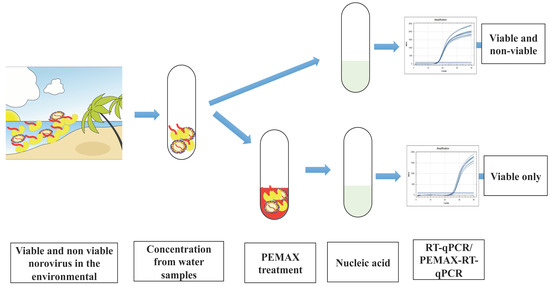
:1. Introduction
2. Materials and Methodology
2.1. Viral Stock Suspensions
2.2. Recovery of Noroviruses from Wastewater and Seawater
2.3. Heat Inactivation of Murine Norovirus and Norovirus
2.4. Optimisation of PEMAXTM Concentration
2.5. Evaluation of PEMAXTM-RT-qPCR for the Selective Detection of Infectious Murine Norovirus
2.6. Evaluation of PEMAXTM-RT-qPCR for the Selective Detection of Infectious Norovirus
2.7. Application of PEMAXTM-RT-qPCR for Detection of Infectious Norovirus from Environmental Samples
2.8. Nucleic Acid Extraction and Virus Detection
2.9. Statistical Analysis
3. Results
3.1. Optimization of PEMAXTM Concentration
3.2. Evaluation of PEMAXTM-RT-qPCR for the Selective Detection of Infectious Murine Norovirus
3.3. Evaluation of PEMAXTM-RT-qPCR for the Selective Detection of Infectious Murine Norovirus
3.4. Application of PEMAXTM-RT-qPCR for Detection of Infectious Norovirus from Environmental Samples
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Ethical Statement
References
- Hewitt, J.; Bell, D.; Simmons, G.C.; Rivera-Aban, M.; Wolf, S.; Greening, G.E. Gastroenteritis outbreak caused by waterborne norovirus at a New Zealand ski resort. Appl. Environ. Microbiol. 2007, 73, 7853–7857. [Google Scholar] [CrossRef] [PubMed]
- Le Guyader, F.S.; Parnaudeau, S.; Schaeffer, J.; Bosch, A.; Loisy, F.; Pommepuy, M.; Atmar, R.L. Detection and quantification of noroviruses in shellfish. Appl. Environ. Microbiol. 2009, 75, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Cangelosi, A.G. , Meschke, S.J. Dead or alive: Molecular assessment of microbial viability. Appl. Environ. Microbiol. 2014, 80, 5881–5891. [Google Scholar] [CrossRef] [PubMed]
- Murakami, K.; Kurihara, C.; Oka, T.; Shimoike, T.; Fujii, Y.; Takai-Todaka, R.; Park, Y.; Wakita, T.; Matsuda, T.; Hokari, R.; et al. Norovirus binding to intestinal epithelial cells is independent of histo-blood group antigens. PLoS ONE 2013, 8, e66534. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; Farkas, T.; Zhong, W.; Tan, M.; Thornton, S.; Morrow, A.L.; Jiang, X. Norovirus and histo-blood group antigens: Demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J. Virol. 2005, 79, 6714–6722. [Google Scholar] [CrossRef] [PubMed]
- Ettayebi, K.; Crawford, S.E.; Murakami, K.; Broughman, J.R.; Karandikar, U.; Tenge, V.R.; Neill, F.H.; Blutt, S.E.; Zeng, X.; Qu, L.; et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 2016, 353, 1387–1393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, M.K.; Watanabe, M.; Zhu, S.; Graves, C.L.; Keyes, L.R.; Grau, K.R.; Gonzalez-Hernandez, M.B.; Iovine, N.M.; Wobus, C.E.; Vinje, J.; et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 2014, 346, 755–759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karim, M.R.; Fout, G.S.; Johnson, C.H.; White, K.M.; Parshionikar, S.U. Propidium monoazide reverse transcriptase PCR and RT-qPCR for detecting infectious enterovirus and norovirus. J. Virol. Methods. 2015, 219, 51–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Randazzo, W.; Lopez-Galvez, F.; Allende, A.; Aznar, R.; Sanchez, G. Evaluation of viability PCR performance for assessing norovirus infectivity in fresh-cut vegetables and irrigation water. Int. J. Food Microbiol. 2016, 229, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, W.; Khezri, M.; Ollivier, J.; Le Guyader, F.C.; Rodríguez-Díaz, J.; Aznar, R.; Sánchez, G. Optimization of PMAxx pretreatment to distinguish between human norovirus with intact and altered capsids in shellfish and sewage samples. Int. J. Food Microbiol. 2018, 266, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Prevost, B.; Goulet, M.; Lucas, F.S.; Joyeux, M.; Moulin, L.; Wurtzer, S. Viral persistence in surface and drinking water: Suitability of PCR pre-treatment with intercalating dyes. Water Res. 2016, 91, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Huang, R.; Chen, H. Evaluation of assays to quantify infectious human norovirus for heat and high-pressure inactivation studies using Tulane virus. Food Environ. Virol. 2017, 9, 314–325. [Google Scholar] [CrossRef] [PubMed]
- Leifels, M.; Jurzik, L.; Wilhelm, M.; Hamza, I.A. Use of ethidium monoazide and propidium monoazide to determine viral infectivity upon inactivation by heat, UV-exposure and chlorine. Int. J. Hyg. Environ. Health 2015, 218, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Sossa, K.E.; Camper, A.K. Molecular monitoring of disinfection efficacy using propidium monoazide in combination with quantitative PCR. J. Microbiol. Methods 2007, 70, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Cheung, C.Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Codony, F.; Agusti, G.; Allue-Guardia, A. Cell membrane integrity and distinguishing between active and inactive cells as a means of improving viability PCR. Mol. Cell Probes 2015, 29, 190–192. [Google Scholar] [CrossRef] [PubMed]
- Wobus, C.E.; Karst, S.M.; Thackray, L.B.; Chang, K.O.; Sosnovtsev, S.V.; Belliot, G.; Krug, A.; Mackenzie, J.M.; Green, K.Y.; Virgin, H.W. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004, 2, e432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewitt, J.; Rivera-Aban, M.; Greening, G.E. Evaluation of murine norovirus as a surrogate for human norovirus and hepatitis A virus in heat inactivation studies. J. Appl. Microbiol. 2009, 107, 65–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewitt, J.; Leonard, M.; Greening, G.E.; Lewis, G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011, 45, 6267–6276. [Google Scholar] [CrossRef] [PubMed]
- Gyawali, P.; Sidhu, J.P.S.; Ahmed, W.; Jagals, P.; Toze, S. Rapid concentration and sensitive detection of hookworm ova from wastewater matrices using a real-time PCR method. Exp. Parasitol. 2015, 159, 5–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kageyama, T.; Kojima, S.; Shinohara, M.; Uchida, K.; Fukushi, S.; Hoshino, F.B.; Takeda, N.; Katayama, K. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J. Clin. Microbiol. 2003, 41, 1548–1557. [Google Scholar] [CrossRef] [PubMed]
- Templeton, M.R.; Andrews, R.C.; Hofmann, R. Particle-associated viruses in water: Impacts on disinfection processes. Crit. Rev. Environ. Sci. Technol. 2008, 38, 137–164. [Google Scholar] [CrossRef]
- Charles, K.J.; Souter, F.C.; Baker, D.L.; Davies, C.M.; Schijven, J.F.; Roser, D.J.; Deere, D.A.; Priscott, P.K.; Ashbolt, N.J. Fate and transport of viruses during sewage treatment in a mound system. Water Res. 2008, 42, 3047–3056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassard, F.; Sharp, J.H.; Taft, H.; LeVay, L.; Harris, J.P.; McDonald, J.E.; Tuson, K.; Wilson, J.; Jones, D.L.; Malham, S.K. Critical review on the public health impact of norovirus contamination in shellfish and the environment: A UK perspective. Food Environ. Virol. 2017, 9, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Ko, G. Using propidium monoazide to distinguish between viable and nonviable bacteria, MS2 and murine norovirus. Lett. Appl. Microbiol. 2012, 55, 182–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Seo, D.J.; Seo, J.; Oh, H.; Jeon, S.B.; Ha, S.D.; Myoung, J.; Choi, I.S.; Choi, C. Detection of viable murine norovirus using the plaque assay and propidium-monoazide-combined real-time reverse transcription-polymerase chain reaction. J. Virol. Methods 2015, 221, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Millard, J.; Appleton, H.; Parry, J.V. Studies on heat inactivation of hepatitis A virus with special reference to shellfish. Epidemiol. Infect. 1987, 98, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, H. Evaluation of the porcine gastric mucin binding assay for high-pressure-inactivation studies using murine norovirus and tulane virus. Appl. Environ. Microbiol. 2015, 81, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Parshionikar, S.; Laseke, I.; Fout, G.S. Use of propidium monoazide in reverse transcriptase PCR to distinguish between infectious and noninfectious enteric viruses in water samples. Appl. Environ. Microbiol. 2010, 76, 4318–4326. [Google Scholar] [CrossRef] [PubMed]
- Baert, L.; Wobus, C.E.; Van Coillie, E.; Thackray, L.B.; Debevere, J.; Uyttendaele, M. Detection of murine norovirus 1 by using plaque assay, transfection assay, and real-time reverse transcription-PCR before and after heat exposure. Appl. Environ. Microbiol. 2008, 74, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Bobay, B.G.; Mertens, B.; Jaykus, L.A. Human norovirus aptamer exhibits high degree of target conformation dependent binding similar to that of receptors and discriminates particle functionality. mSphere 2016, 1, e00298-16. [Google Scholar] [CrossRef] [PubMed]
- Samandoulgou, I.; Hammami, R.; Rayas, R.M.; Fliss, I.; Jean, J. Stability of secondary and tertiary structures of virus like particles representing noroviruses: Effects of pH, ionic strength, and temperature and implications for adhesion to surfaces. Appl. Environ. Microbiol. 2015, 81, 7680–7686. [Google Scholar] [CrossRef] [PubMed]
- Fuster, N.; Pinto, R.M.; Fuentes, C.; Beguiristain, N.; Bosch, A.; Guix, S. Propidium monoazide RTqPCR assays for the assessment of hepatitis A inactivation and for a better estimation of the health risk of contaminated waters. Water Res. 2016, 101, 226–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Dilutions | Relative log10 Reduction in Norovirus Concentration | |||
|---|---|---|---|---|
| GI.3 | GII.4 | |||
| Original | Heat-Inactivated | Original | Heat-Inactivated | |
| 100 | 0.11 ± 0.02 | 0.91 ± 0.16 | 0.05 ± 0.08 | 1.12 ± 0.13 |
| 10−1 | 0.05 ± 0.25 | 0.47 ± 0.12 | 0.06 ± 0.01 | 2.03 ± 0.08 |
| 10−2 | 0.17 ± 0.11 | 0.34 ± 0.21 | 0.13 ± 0.03 | 0.63 ± 0.07 |
| 10−3 | 0.17 ± 0.28 | 0.43 ± 0.13 | 0.15 ± 0.01 | 0.53 ± 0.03 |
| 10−4 | 0.05 ± 0.04 | 0.98 ± 0.35 | 0.25 ± 0.09 | 2.26 ± 0.02 |
| Sample Description | Sample ID | Norovirus GI | Norovirus GII | ||||
|---|---|---|---|---|---|---|---|
| qPCR CT Values (Mean ± SD) | ΔΔ CT | qPCR CT Values (Mean ± SD) | ΔΔ CT | ||||
| (−) PEMAXTM | (+) PEMAXTM | (−) PEMAXTM | (+) PEMAXTM | ||||
| Influent wastewater | I1 | 34.9 ± 0.24 | 36.6 ± 0.73 | −1.65 | 33.2 ± 0.61 | 33.8 ± 0.36 | −0.53 |
| I2 | 36.5 ± 0.60 | 38.6 ± 0.15 | −2.14 | 28.1 ± 0.04 | 29.5 ± 0.07 | −1.42 | |
| I3 | 34.9 ± 0.28 | 35.2 ± 0.23 | −0.30 | 27.2 ± 0.07 | 27.4 ± 0.12 | −0.17 | |
| I4 | ND | ND | NA | 40.6 ± 1.01 | 40.9 ± 0.67 | −0.22 | |
| I5 | 37.5 ± 0.10 | 39.8 ± 0.23 | −2.23 | 31.5 ± 0.07 | 32.6 ± 0.14 | −1.15 | |
| I6 | 30.4 ± 0.36 | 31.9 ± 0.10 | −1.42 | 31.4 ± 0.01 | 32.7 ± 0.23 | −1.29 | |
| Effluent wastewater | E1 | 39.6 ± 0.32 | 40.4 ± 0.10 | −0.76 | 37.2 ± 0.26 | 39.3 ± 0.98 | −2.05 |
| E2 | 33.1 ± 0.26 | 34.7 ± 0.06 | −1.63 | 28.3 ± 0.09 | 29.3 ± 0.12 | −0.94 | |
| Seawater | S1 | 40.8 ± 0.59 | ND | NA | 37.8 ± 0.36 | 39.0 ± 0.04 | −1.12 |
| S2 | 40.2 ± 1.05 | ND | NA | 37.4 ± 0.39 | ND | NA | |
| Sample Description | Sample ID | Percentage of Infectious Norovirus Detected | |
|---|---|---|---|
| GI | GII | ||
| Influent wastewater | I1 | 32 | 59 |
| I2 | 23 | 37 | |
| I3 | 81 | 89 | |
| I4 | NA | 86 | |
| I5 | 21 | 45 | |
| I6 | 38 | 41 | |
| Effluent wastewater | E1 | 59 | 24 |
| E2 | 32 | 52 | |
| Seawater | S1 | 0 | 44 |
| S2 | 0 | 0 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gyawali, P.; Hewitt, J. Detection of Infectious Noroviruses from Wastewater and Seawater Using PEMAXTM Treatment Combined with RT-qPCR. Water 2018, 10, 841. https://doi.org/10.3390/w10070841
Gyawali P, Hewitt J. Detection of Infectious Noroviruses from Wastewater and Seawater Using PEMAXTM Treatment Combined with RT-qPCR. Water. 2018; 10(7):841. https://doi.org/10.3390/w10070841
Chicago/Turabian StyleGyawali, Pradip, and Joanne Hewitt. 2018. "Detection of Infectious Noroviruses from Wastewater and Seawater Using PEMAXTM Treatment Combined with RT-qPCR" Water 10, no. 7: 841. https://doi.org/10.3390/w10070841